Span Categorization New: 1.11
A common task in applied NLP is extracting spans of texts from documents,
including longer phrases or nested expressions. Prodigy lets you label
potentially overlapping and nested spans and create training data for models and
components like spaCy’s
SpanCategorizer.
Named Entity Recognition vs. Span Categorization
Named entity recognition is very effective for proper nouns and self-contained expressions like person names or products, because it predicts single-token-based tags and takes advantage of clear span boundaries. However, it’s usually not the right tool for other types of phrases, sentence fragments and overlapping spans with less consistent boundaries and mixed lengths. For use cases like this, you typically want to choose a model that predicts scores and labels over a selection of spans. We call this type of component a span categorizer.
| Named Entity Recognition | Span Categorization |
|---|---|
| spans are non-overlapping syntactic units like proper nouns (e.g. persons, organizations, products) | spans are potentially overlapping units like noun phrases or sentence fragments |
model predicts single token-based tags like B-PERSON with one tag per token | model predicts scores and labels for suggested spans |
| takes advantage of clear token boundaries | less sensitive to exact token boundaries |
Before you get started on your NLP project, it can often be very helpful to take a step back and think about the type of information you really need for your downstream application and which NLP component is the best fit to capture this information in your data and annotate it consistently. In some cases, it can make more sense to opt for a text classifier instead of a span categorizer, which will predict one or more labels over the whole text and lets you capture signals from multiple places across the text, instead of attaching them to specific spans of text.
Fully manual annotation
To get started with manual span annotation, all you need is a file with raw
input text you want to annotate and a
spaCy pipeline for tokenization (so the web app
knows what a word is and can allow more efficient highlighting). The following
command will start the web server with the spans.manual recipe, stream in
texts from journal_papers.jsonl and provides the label options FACTOR,
CONDITION, METHOD and EFFECT. The spans_manual UI will then let
you annotate arbitrary and potentially overlapping spans that are displayed
below the tokens.
Example
prodigy spans.manual covid_articles blank:en ./journal_papers.jsonl --label FACTOR,CONDITION,METHOD,EFFECT
To make annotation faster, the selection automatically “snaps” to token boundaries and you can double-click on a single token to select it. Hovering a span label will highlight all tokens it refers to. To remove a span, you can click on its label.
Working with patterns
Just like ner.manual, the spans.manual workflow supports providing
a match patterns file. Match patterns are
typically provided as a JSONL (newline-delimited JSON) file and can be used to
pre-highlight spans for faster and more efficient annotation. Since spans can
overlap, the found matches are not filtered and all matched spans will be
pre-highlighted in the UI.
Example
prodigy spans.manual covid_articles blank:en ./journal_papers.jsonl --label CONDITION --patterns ./condition_patterns.jsonl
Working with suggester functions
spaCy’s SpanCategorizer works by using
a suggester function to suggest spans that the model will then predict
labels for. A common strategy is to use ngrams of certain sizes, e.g. all
possible spans of lengths 1 to 10. You can also implement more sophisticated
suggesters, for instance to consider all noun phrases in Doc.noun_chunks, or
only certain spans defined by
match patterns.
Letting Prodigy infer the best-matching ngram suggester
During development, it’s often helpful to just start annotating and get a sense
for the data and the types of spans you’re looking for. If you don’t provide a
config with a suggester during training, Prodigy will use the collected
annotations to infer the best matching ngram suggester based on the available
spans. For example, if your shortest span is 1 token and your longest span is 5
tokens, the created ngram suggester will cover a range of 1 to 5.
Setting the --verbose flag on train will output a detailed breakdown of
the spans found in the data. This is also useful for discovering common patterns
and outliers, or even potential mistakes. As always with machine learning, it’s
very important that your training data is representative of what your model
will see at runtime.
Example (excerpt)
prodigy train --spancat covid_articles,eval:covid_articles_eval --verbose ======================== Generating Prodigy config ======================== ℹ Auto-generating config with spaCy Using 'spacy.ngram_range_suggester.v1' for 'spancat' with sizes 1 to 5 (inferred from data) Sizes: [1, 2, 3, 4, 5] - 1 (298 spans) - 2 (96 spans) - 3 (15 spans) - 4 (1 spans) - 5 (8 spans) ✔ Generated training config
The inferred suggester sizes will be added to your config.cfg that’s exported
with the trained pipeline and generated when you run data-to-spacy. Once
you got a better sense for the types of spans and span sizes you’re expecting,
it can often be useful to run different experiments with different suggesters
and compare the results.
config.cfg (excerpt)[components.spancat.suggester]
@misc = "spacy.ngram_range_suggester.v1"
min_size = 1
max_size = 5
Annotating with suggester functions
Prodigy lets you provide a suggester function when you annotate with
spans.manual, so you can make sure that the spans you annotate match the
suggested spans you want to train on. This way, you’ll never end up with
annotations describing spans that aren’t ever suggested and that the model
couldn’t learn from. The suggester is run via the
validate_answer callback every time an
annotator submits an answer. If an annotated span doesn’t match the suggested
spans, you’ll see an alert in the UI with more information, and you’ll only
be able to submit the annotation if the validation passes.
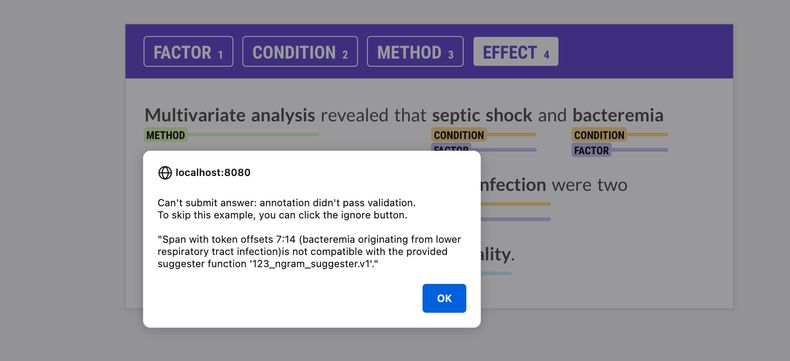
The name of the suggester function can be provided via --suggester. You can
use Prodigy’s -F argument to load in a Python file containing the registered
suggester function, which should be registered in spaCy’s
@misc registry. If your suggester
requires annotations from other components you can use the --use_annotations
argument, which uses the defined spaCy model to process the text and to save the
annotations.
Example with suggester validation
prodigy spans.manual covid_articles blank:en ./journal_papers.jsonl --label CONDITION --suggester 123_ngram_suggester.v1 -F ./suggester.py --use_annotations
suggester.pyfrom spacy import registry
from spacy.pipeline.spancat import build_ngram_suggester
@registry.misc("123_ngram_suggester.v1")
def custom_ngram_suggester():
return build_ngram_suggester(sizes=[1, 2, 3]) # all ngrams of size 1, 2 and 3
When you train your span categorizer, you can define the suggester function to
use in your config.cfg. If
you’ve validated your annotations against the same suggester, you’ll know that
all examples you’re training from match the suggested spans the model will see.
config.cfg (excerpt)[components.spancat.suggester]
@misc = "spacy.ngram_suggester.v1"
sizes = [1, 2, 3]
Training a span categorizer
Training a spaCy pipeline
The easiest way to run a quick experiment is to use Prodigy’s train
workflow, which is a thin wrapper around spaCy’s
training API. The --spancat argument lets
you provide one or more datasets to train from. You can optionally provide
dedicated evaluation sets using the eval: prefix. If no evaluation set is
provided, a percentage of examples is held back for evaluation. If no custom
config is specified, Prodigy will take care of
inferring the best-matching suggester to use based on
the data.
Example
prodigy train ./model --spancat covid_articles,eval:covid_articles_eval
Instead of training with Prodigy, you can use the data-to-spacy command
to export a ready-to-use corpus and config for training with spaCy directly:
Example (excerpt)
prodigy data-to-spacy ./corpus --spancat covid_articles To use this data for training with spaCy, you can run: python -m spacy train ./corpus/config.cfg --paths.train ./corpus/train.spacy --paths.dev ./corpus/dev.spacy
Predicted span annotations are available as a list of
Span objects via the
Doc.spans, a dictionary of span groups. This
allows you to store multiple collections of spans from different sources on the
Doc. By default, annotations exported with Prodigy will use the key "sc" for
”span categorization”:
Usage in spaCynlp = spacy.load("./model")
doc = nlp("Septic shock and bacteremia were two independent risk factors.")
print(doc.spans)
# {'sc': [Septic shock, Septic shock, bacteremia, bacteremia]}
print([(span.text, span.label_) for span in doc.spans["sc"]])
# [('Septic shock', 'FACTOR'), ('Septic shock', 'CONDITION'), ('bacteremia', 'FACTOR'), ('bacteremia', 'CONDITION')]
Exporting your annotations
The db-out exports annotations in a straightforward JSONL format. Spans
are provided as a list of "spans" with their respective start and end
character offsets, and the entity label. This should make it easy to convert and
use it to train any model. If you annotated using a manual interface, the data
will also include a "tokens" property so you can restore the tokenization.
Example
prodigy db-out covid_articles > ./annotations.jsonl